FIRST, DOWNLOAD & OPEN THE PDB STRUCTURE 2JFF.
1) Then, expand the Molecule Tree View tool of 2JFF.
2) Left-click on "1440 LK2". Below image shown after clicking "1440 LK2".
3) Select Edit & click Hide Unselected menu.
4) Then, select View and click Center Moecule in Window menu.
5) Right-click on "1440 LK2". Then, add Hydrogen by clicking Hydrogen menu.
6) Select the "Make a Ligand Group from this Residue" option.
7) The Group folder shown "1 LK2".
8) Copy and Paste the "1440 LK2".
9) Select "Make a Ligand Group from this Residue".
10 ) There are two Ligands in the Groups folder named "1 LK2" and "2 LK2".
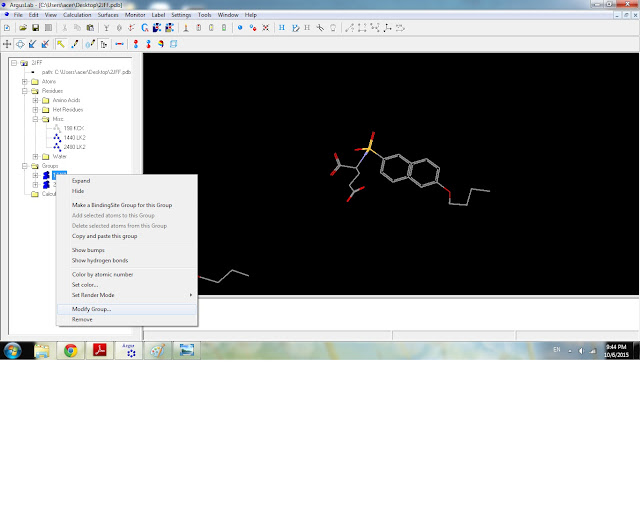
11) Rename the two Ligand groups. First, click "Modify Group".
12) "1 LK2" changes to "ligand-xray" while "2 LK2" changes to "ligand".
13) Here is the image after renaming the ligand group.
14) To more easily distinguish the two ligand in the graphics window, you can do a number of different things.
15) Right click the ''ligand-xray" & select the "Set Render Mode". Then, choose "Cylinder med" menu.
16) Here is the image.
17) Click the "Make a BindingSite Group for this Group" menu.
18) HERE is the image after rightclick "Make a BindingSite Group for this Group".
19) Now, try to dock a ligand.
19) It will show like this.
20) Select the ligand to dock in the "Ligand" drop-box. Then, click "start" menu
21) The image shown after clicking "Start".
22) There a listing of the docking settings and several "poses".
23) Select both the ligand and ligand-xray groups by holding down the "Ctrl" key and left-clicking on both groups in the Groups folder. Both groups should be highlighted on the screen.
24) Now, select ""Calc RMSD position between two similar Groups".
25) RMSD value should be less than approximately 2.5 Angstroms.



















nice thanks a lot but could you explain why we take 2 similar ligand? why not (receptor and ligand) ? and i couldn't find the last 2 steps 24 and 25. could you explain more about the results pleaaaase.
ReplyDelete